Research to be presented at the 2021 American Academy of Pediatrics National Conference & Exhibition is examining proteins in the saliva of patients with COVID-19 infection
ITASCA, IL – An ongoing study is looking at the relationship between cytokines in saliva and COVID-19 infection to help predict the severity of infection. In a preliminary analysis of saliva samples from 150 children, the researchers found that levels of two cytokines were higher in those with severe COVID-19 compared to those without severe infection.
The study abstract, “Severity Predictors Integrating Salivary Transcriptomics and Proteomics with Multineural Network Intelligence in SARS-CoV2 infection in Children; SPITS-MISC,” to be presented at the virtual American Academy of Pediatrics 2021 National Conference & Exhibition, is looking at cytokines and microRNAs (non-coding RNAs) in saliva in children. These biomarkers may control the inflammation in the body once infected with the virus and help determine the seriousness of the infection.
“Using saliva to predict severity of the infection is non-invasive and painless. If proven to be effective saliva may be a game changer in children in whom obtaining blood is both difficult and distressing. Additionally, early recognition of the severity of COVID-19 can help clinicians institute timely and appropriate treatment which may help improve outcomes.” said Dr. Usha Sethuraman of Central Michigan University and DMC Children’s Hospital of Michigan, an author of the study.
The majority of children with COVID-19 infection have had mild illness, but some children have developed severe complications such as respiratory failure or inflammation of the heart, the authors note. Cytokines are proteins found in blood and saliva that may be produced in response to an infection. Studies in adults have shown that certain cytokines are elevated in the blood of patients with COVID-19 and may predict how severe the illness will be.
The goal of the study is to be able to identify children at risk for severe disease by integrating these biomarkers and social determinants of health using artificial intelligence. Dr. Sethuraman, Dr. Steven D Hicks of Penn State College of Medicine, and Dr. Dongxiao Zhu of Wayne State University are obtaining saliva samples from 400 children ages 18 and younger with COVID-19 infection who seek emergency medical care at two children’s hospitals: Children’s Hospital of Michigan and UPMC Children’s Hospital of Pittsburgh. Analysis of saliva samples is being performed at Penn State College of Medicine and model development using artificial intelligence is being performed at Wayne State University.
In addition to finding that levels of two cytokines (MIG and CXCL-10) in the preliminary analysis were higher in those with severe COVID-19 compared to those without severe infection, dozens of microRNA levels were found to be altered, with the majority of them being significantly lower in the saliva of children with severe infection. Ongoing analysis will seek to validate these results and confirm the importance of saliva cytokines and microRNAs, combined with social factors, including where children live.
The study was supported by a grant from the Eunice Kennedy Shriver National Institute of Child Health and Human Development through the National Institutes of Health’s Rapid Acceleration of Diagnostics program (1R61HD105610).
Dr. Usha Sethuraman will present the study on Oct. 8 at 3.01p.m. CT.
To request an interview, journalists may contact Kate Worster at [email protected], Aaron Mills at [email protected], Jason Barczy at [email protected], or Dr. Usha Sethuraman at [email protected].
Please note: Only the abstract is being presented at the meeting. In some cases, the researcher may have more data available to share with media or may be preparing a longer article for submission to a journal.
# # #
The American Academy of Pediatrics is an organization of 67,000 primary care pediatricians, pediatric medical subspecialists and pediatric surgical specialists dedicated to the health, safety and well-being of infants, children, adolescents and young adults. For more information, visit www.aap.org.
Program Name:
Abstract Title: MicroRNA Profiles in Saliva of Children with and Without Severe Disease Due to Severe Acute
Usha Sethuraman
Detroit, MI, United States
(313) 745-5260
Monday, October 11, 2021: 11:15 AM –
The majority of children with exposure to SARS-CoV-2 virus have mild disease. However severe diseases such as Multisystem inflammatory syndrome (MISC) and pneumonia do occur in children. Currently, there are no established biomarkers that can predict progression to severe disease in children exposed to the virus. MicroRNAs (miRNAs) are non-coding RNAs that can be found in saliva and are thought to play a role in the regulation of inflammation following an infection. Our objective was to compare the miRNA profile in saliva of children with or without severe disease due to SARS-CoV-2 infection.
This prospective observational study was supported by the National Institutes of Health (NIH) RADx Program. Children ≤ 18 years of age presenting to two tertiary care children’s hospitals with symptoms of SARS-CoV-2 infection (confirmed by PCR test, serology or epidemiological link) were enrolled between 03/29/2021 and 04/ 30/2021. Severe infection was defined as any of the following within 30 days of testing: MISC or Kawasaki disease diagnosis, requirement for oxygen > 2L, inotropes, mechanical ventilation or ECMO, or the occurrence of death. Informed consent and a saliva swab were obtained at the time of SARS-CoV-2 diagnosis (DNA Genotek, Ottowa Canada), and RNA was extracted (Qiagen, Germantown, MD). Small RNA species (<50 base pairs) were interrogated via shotgun sequencing (HiSeq 2500, Illumina, San Diego, CA) and miRNAs were quantified through alignment to the human genome (GRCh38). RNA features with sparse counts (<10 in 90% of samples) were filtered, and the data was quantile normalized and mean-center scaled. Salivary miRNA levels were compared between those with severe and non-severe SARS-CoV-2 infection using Wilcoxon tests with Benjamini Hochberg multiple testing corrections. In addition, a logistic regression analysis was used to identify miRNA pairs that could best discriminate severe cases based on a Monte Carlo 100-fold cross-validated area under receiver operating characteristic curve (AUROC).
Samples from 33 children were analyzed. Median age was 3 (3, 10) years and 54.5 % were males. Of the total, 29 were RT PCR positive, 4 had a positive serology and 6 children had severe infection. Seven miRNAs displayed significant differences (Fold change >2, FDR adjusted p < 0.1) among children with severe SARS-CoV-2 infection (Table). All seven miRNAs were up-regulated in severe SARS-CoV-2 cases. A logistic regression using a single ratio of miR-296-5p/miR-378j yielded 1.0 AUROC for differentiating children with severe infection (Figure).
In this interim analysis of salivary miRNA in childhood SARS-CoV-2 infection, we found a differential expression of 7 salivary miRNAs in children with severe infection. Ongoing work will seek to validate these findings and explore the role of miRNA in predicting severe SARS-CoV-2 infection in children.
Table
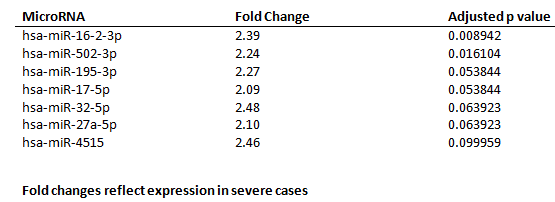
Salivary microRNAs with significant differences between children with severe and non-severe SARS-CoV-2 infection
Figure.
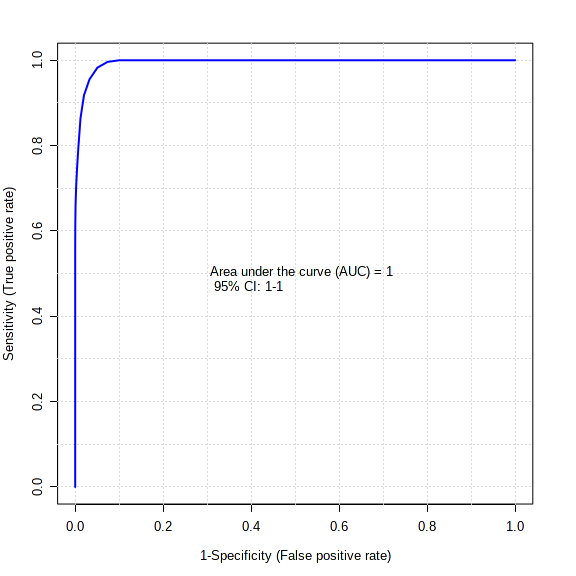
Receiver operating characteristic curve and box plot displaying the complete differentiation of severe and non- severe SARS-CoV-2 cases using a ratio of miR-296-5p and miR-378j levels in saliva.